Introducing openstatsware and the R Package {mmrm}
China-R Conference
2023-11-28
Acknowledgments
Authors:
- Brian Matthew Lang (MSD)
- Christian Stock (BI)
- Craig Gower-Page (Roche)
- Dan James (AstraZeneca)
- Daniel Leibovitz (Roche)
- Daniel Sabanes Bove (Roche, lead)
- Daniel Sjoberg (Roche)
- Doug Kelkhoff (Roche)
- Julia Dedic (Roche)
- Jonathan Sidi (PinPoint Strategies)
- Kevin Kunzmann (BI)
- Ya Wang (Gilead)
Acknowledgments
Thanks to:
- Ben Bolker (McMaster University)
- Davide Garolini (Roche)
- Dinakar Kulkarni (Roche)
- Gonzalo Duran Pacheco (Roche)
- Software Engineering working group (
openstatsware)
Agenda
- Introduction and Overview of
openstatsware - Mixed Models for Repeated Measures - Why is it a Problem?
- MMRM Package
- Why this is not “yet another package”
- Comparing MMRM R Package to other Implementations
- Get Started with
{mmrm}!
Context: Software Engineering in Biostatistics
- Open-source software has gained increasing popularity in Biostatistics
- Pros: rapid uptake of novel statistical methods, unprecedented opportunities for collaboration
- Cons: variability in software quality
- Developing high-quality software is critical
Software Engineering working group (openstatsware)
- Official working group of the American Statistical Association (ASA) Biopharmaceutical Section
- Formed on 19 August 2022
- Cross-industry collaboration (59 members from 38 organizations)
- Full name: Software Engineering Working Group
- Short name:
openstatsware - Home page at rconsortium.github.io/asa-biop-swe-wg
- We welcome new members to join!
Working Group Objectives
- Primary
- Engineer R packages that implement important statistical methods
- to fill in gaps in the open-source statistical software landscape
- focusing on what is needed for biopharmaceutical applications
- Engineer R packages that implement important statistical methods
- Secondary
- Develop and disseminate best practices for engineering high-quality open-source statistical software
- By actively doing the statistical engineering work together, we align on best practices and can communicate these to others
- Develop and disseminate best practices for engineering high-quality open-source statistical software
First Product of openstatsware: mmrm
- MMRM is a popular choice for analyzing longitudinal continuous outcomes in randomized clinical trials
- No great R package
- Initially thought that the MMRM problem was solved by using
lme4withlmerTest, learned that this approach failed on large data sets (slow, did not converge) nlmedoes not give Satterthwaite adjusted degrees of freedom, has convergence issues, and withemmeansit is only approximate- Next we tried to extend
glmmTMBto calculate Satterthwaite adjusted degrees of freedom, but it did not work
- Initially thought that the MMRM problem was solved by using
Idea
- We only want to fit a fixed effects model with a structured covariance matrix for each subject
- The idea is then to use the Template Model Builder (
TMB) directly- as it is also underlying
glmmTMB - but code the exact model we want
- as it is also underlying
- We do this by implementing the log-likelihood in
C++using theTMBprovided libraries - Provide an R solution that
- has fast convergence times
- generates estimates closest to
SAS
Advantages of TMB
- Fast
C++framework for defining objective functions (Rcppwould have been alternative interface) - Automatic differentiation of the log-likelihood as a function of the variance parameters
- We get the gradient and Hessian exactly and without additional coding
- Syntactic sugars to allow simple matrix calculations or operations like R
- This can be used from the R side with the
TMBinterface and plugged into optimizers
Why it’s not just another package
- Ongoing maintenance and support from the pharmaceutical industry
- 5 companies being involved in the development, on track to become standard package
- Development using best practices as show case for high quality package
- Thorough unit and integration tests (also comparing with SAS results) to ensure accurate results
Highlighted Features of mmrm
- Covariance structures for the dependent observations:
- Unstructured, Toeplitz, AR1, compound symmetry, ante-dependence, spatial exponential
- Allows group specific covariance estimates and weights
- Hypothesis Testing:
emmeansinterface for least square means- Satterthwaite and Kenward-Roger adjustments
- Robust sandwich estimator for covariance
- Integrations and extentions
tidymodelsbuiltin parsnip engine and recipes for streamlined model fitting workflowsteal,tern,rtablesintegration for post processing and reporting- Support conditional mean prediction and simulation
- Also used in
rbmifor conditional mean imputation!
Comparison with Other Software
To run an MMRM model in SAS it is recommended to use either the PROC MIXED or PROC GLMMIX procedures.
Less model assumptions are applied in
PROC MIXED, primarily how one treats missingness.We will compares the
PROC MIXEDprocedure to the{mmrm}package in the following attributes:
- Documentation
- Unit Testing
- Covariance structures
- Covariance Estimators
- Degrees of Freedom
- Contrasts
Documentation
Both languages have online documentation of the technical details of the estimation and degrees of freedom methods and the different covariance structures available.
{mmrm}
PROC MIXED
Unit Testing
- Unit tests in
{mmrm}are transparent, compared toPROC MIXED. - Uses the
testthatframework withcovrto communicate the testing coverage. - Unit tests can be found in the GitHub repository under ./tests.
Note
The integration tests in {mmrm} are set to a tolerance of 10e^-3 when compared to SAS outputs.
Covariance structures
- SAS has 23 non-spatial covariance structures, while mmrm has 10.
- 9 structures intersect with SAS
- Ante-dependence (homogeneous) is only in the mmrm package.
- SAS has 14 spatial covariance structures compared to the spatial exponential one available in mmrm.
Tip
For users that need more structures, {mmrm} is easily extensible - let us know via feature requests in the GitHub repository.

Covariance structures details
| Covariance structures | {mmrm} |
PROC MIXED |
|---|---|---|
| Unstructured (Unweighted/Weighted) | X/X | X/X |
| Toeplitz (hetero/homo) | X/X | X/X |
| Compound symmetry (hetero/homo) | X/X | X/X |
| Auto-regressive (hetero/homo) | X/X | X/X |
| Ante-dependence (hetero/homo) | X/X | X |
| Spatial exponential | X | X |
Covariance Estimators
| Covariance estimators | {mmrm} |
PROC MIXED |
|---|---|---|
| Asymptotic | X | X |
| Kenward-Roger | X | X |
| Empirical | X | X |
| Jackknife | X | |
| Bias-Reduced Linearization | X |
Degrees of Freedom Methods
| Method | {mmrm} |
PROC MIXED |
|---|---|---|
| Contain | X* | X |
| Between/Within | X* | X |
| Residual | X* | X |
| Satterthwaite | X | X |
| Kenward-Roger ** | X | X |
| Kenward-Roger (Linear) | X | X |
*Available through emmeans, and ongoing work in mmrm.
** Unstructured has differences compared to PROC MIXED because of different parameterization.
Contrasts/LSMEANS
Contrasts and least square (LS) means estimates are available in mmrm using
mmrm::df_1d,mmrm::df_md- S3 method that is compatible with
emmeans - LS means difference can be produced through emmeans (
pairsmethod) - Degrees of freedom method is passed from
mmrmtoemmeans - By default
PROC MIXEDandmmrmdo not adjust for multiplicity, whereasemmeansdoes.
Note
These are comparable to the LSMEANS statement in PROC MIXED.
Benchmarking with other R packages
Other R packages, like nlme, lme4 and glmmTMB can support MMRM analysis. However:
- Covariance structure supported is limited
- Computational efficiency is not satisfying
- Convergence issues(especially for
lme4) - Need other(multiple) packages to support
SatterthwaiteandKenward-Rogerdegrees of freedom, or Sandwich covariance estimator. - Detailed results at the online comparison vignette
Computational Efficiency
mmrm not only supports multiple covariance structure, it also has good efficiency (due to fast implementations in C++)

Differences with SAS
{mmrm} has small difference from SAS
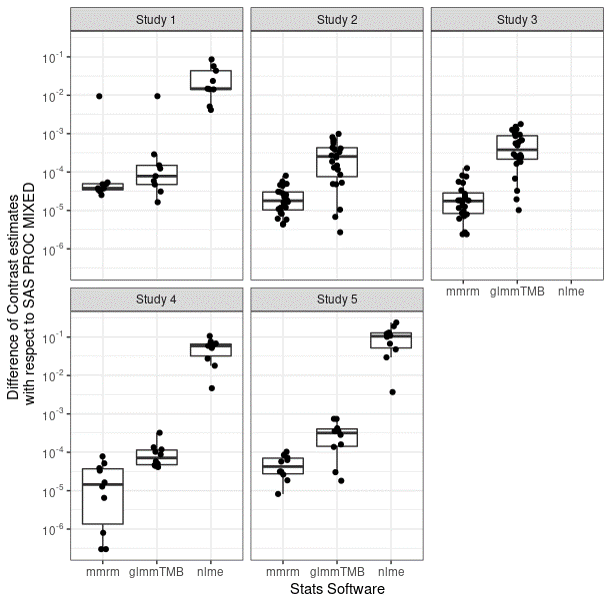
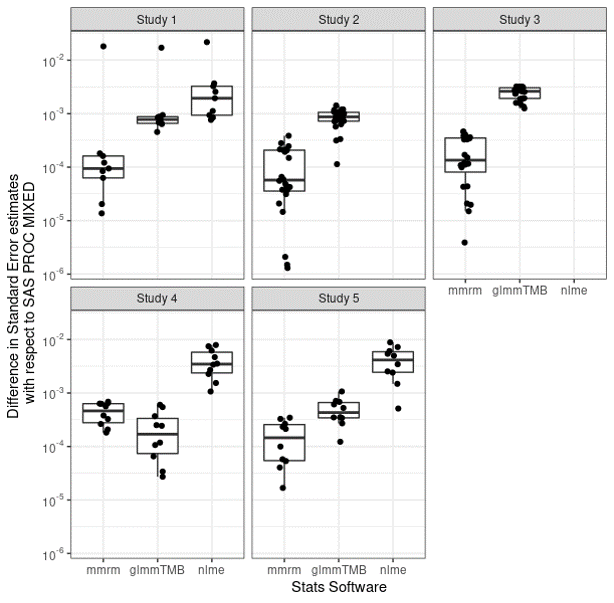
Getting started
mmrmis on CRAN - use this as a starting point:
- Visit openpharma.github.io/mmrm for detailed docs including vignettes
- Consider tern.mmrm for high-level clinical reporting interface, incl. standard tables and graphs
Thank you! Questions?


